JXB Volume 72, Issue 20 – Editor’s choice
JXB VOLUME 72, ISSUE 20 – EDITOR’S CHOICE
This article highlights the following publication:
Differential degradation of RNA species by autophagy-related pathways in Arabidopsis
Daniel Hickl, Franziska Drews, Christopher Girke, David Zimmer, Timo Mühlhaus, Jan Hauth, Karl Nordström, Oliver Trentmann, Ekkehard H. Neuhaus, David Scheuring, Tobias Fehlmann, Andreas Keller, Martin Simon and Torsten Möhlmann
Journal of Experimental Botany, Volume 72, Issue 20, 26 October 2021, Pages 6867–6881, https://doi.org/10.1093/jxb/erab321
Accompanying Insight:
Eating the messenger (RNA): autophagy shapes the cellular RNA landscape
Girishkumar Kumaran and Simon Michaeli
Journal of Experimental Botany, Volume 72, Issue 20, 26 October 2021, Pages 6803–6807, https://doi.org/10.1093/jxb/erab385
Sophisticated recycling system: How plants dispose of waste RNA
The global Covid-19 pandemic and the subsequent breakthrough of broad-scale mRNA vaccine application brought about widespread interest into the fragile nature of RNA molecules. Although decades of intensive research and painstaking design modifications have gone into their development, mRNA-based vaccines are still only stable for several hours at room temperatures and they need to be shipped and stored at ultra-low temperatures, making their use in warm countries and remote areas logistically challenging. The inherent susceptibility of RNA to degradation is presumably not an evolutionary accident but has instead been favoured by natural selection, and rapid enzymatic RNA degradation by RNases is a key regulatory process in gene expression. Only constant RNA turnover to prevent excess accumulation gives cells the freedom to adequately respond to external or internal signals and furthermore, allows for recycling of nucleoside components. The steady-state abundance of cellular RNA thus reflects an equilibrium between synthesis and degradation, and the half-life of most RNA is less than 10 minutes.
In plants only a fraction of RNA degradation takes place within the cytosol whereas the highest RNase activity can be detected in the vacuole, an organelle that is much more prominent in plant compared to animal cells, and that fulfils diverse functions including turgor regulation, storage of minerals and metabolites, and controlled degradation of unwanted cellular components including RNA molecules. Uptake of waste material into the vacuole is facilitated by a process called “autophagy”, in which membrane-bound vesicles form in the cytosol and fuse with the vacuole membrane to release their cargo. In the latest JXB issue, Hickl et al. (2021) explore the role that autophagy plays in RNA decay. RNA can also be delivered to the vacuole via alternative pathways and the contribution of autophagy to vacuolar RNA decomposition has not yet been quantified. Also, the specificity of this process remained uncertain thus far. Therefore, the authors have developed a protocol to isolate and sequence vacuolar RNA at single nucleotide level, in order to compare the cellular and vacuolar RNAome and gain insight into the fate of different RNA species. Whereas messenger RNA (mRNA), transfer RNA (tRNA) and ribosomal RNA (rRNA) are directly involved in protein biosynthesis, small, non-coding RNAs such as miRNA can control gene expression via post-transcriptional or chromatin-dependent gene silencing. All of these RNA types were detected in vacuolar preparations with pre-tRNA and rRNA being the predominant RNA species. Within the vacuole, RNS2 is the most prominent RNase in Arabidopsis, and knocking down this enzyme decreased the vacuolar RNA degradation activity to about 30% compared to wildtype plants. Despite this marked reduction in RNA hydrolysis the average length of vacuolar RNA fragments in the mutant was not significantly different from the wildtype, indicating the existence and activity of additional compensatory vacuolar RNases. To elucidate the role of autophagy in vacuolar RNA degradation, the authors test Arabidopsis mutants that lack a key component of the autophagy process. Their analysis showed that especially chloroplast-encoded RNA related to photosynthesis was markedly reduced in the mutant vacuoles, indicating that RNA delivery to the vacuole via autophagy is indeed specific for certain RNA types and especially important for chloroplast RNA recycling. By contrast, photosynthesis-related transcripts of nuclear origin were increased in the mutant’s vacuole and the authors presume the activity of autophagy-independent delivery pathways to the vacuole for these RNA types.
A surprising finding of the vacuolar RNAome sequencing presented by Hickl et al. was the presence of intact vacuolar miRNA that are presumably protected from degradation. These small RNA fragments play important roles in post-transcriptional gene regulation via RNA silencing. Their accumulation in the vacuole presents a molecular mystery that remains to be investigated in future studies.
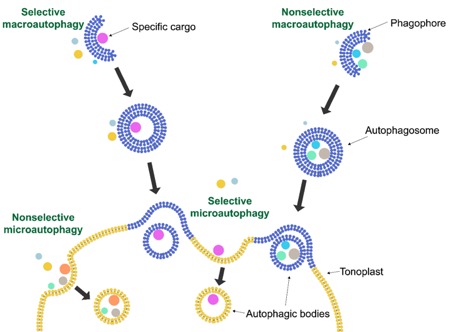
Autophagy in plants describes the uptake of cellular material destined for degradation into the vacuole via membrane-bound vesicles. This process is beautifully explained and illustrated in a recent JXB Insight article by Kumaran and Michaeli (2021), highlighting the findings of Hickl et al. (2021), who investigate the role of autophagy in vacuolar RNA degradation.





