Protecting our plants
Although ‘normal life’ has been grounded by a virus, for plants it is fungal pathogens (and fungal-like oomycetes) that pose the biggest threat. “All five staple crops that provide most of our calories are threatened by devastating fungal pathogens,” says Sarah. It has been estimated that the amount lost of these key crops to fungi and oomycetes alone would be enough to feed 8.5% of today’s population.1 And two major drivers—climate change and rapid fungal evolution—mean this problem is only set to grow larger.
Climate change has already expanded the range of many fungal pathogens, allowing them to move northwards at an alarming rate.2 Sarah and her colleague Dan Bebber have been involved in efforts to document this change using historical records, and to then apply this knowledge to develop a predictive model of how particular fungi respond to climate change. The model was based on experimental data gathered by Helen Fones and PhD student Tom Chaloner that assessed how temperature and/or wetness affected every lifecycle stage of the model fungal pathogen Zymoseptoria tritici.3 This informed the concept of a ‘disease triangle’ that incorporates three facets: environmental conditions, pathogen presence and virulence, and host presence and susceptibility.4 “Using other models, we can now begin to predict more accurately where particular pathogens should be found, and how this is likely to change,” says Sarah. “One of our students even went out to China and successfully found several ‘missing’ fungal crop pathogens that our global model had predicted, even though they had not been reported in the western literature in these regions before.”
Knowing where pathogens are likely to be is of little use unless we can effectively tackle them. Because our major crops are typically grown as vast swathes of monocultures with almost no inherent genetic variability, fungicides play a critical role in controlling fungal pathogens. Yet these have a fundamental weakness in that they typically target only a single site in the pathogen. This allows pathogens to rapidly evolve to become resistant.5 “We urgently need new classes of fungicide that have multiple modes of activity,” says Sarah. “I joined a project led by Gero Steinberg and his team, Sreedhar Kilaru and Martin Schuster, with the help of organic chemist Mark Wood. We used an approach combining plant pathology, fungal cell biology, toxicology, and synthetic chemistry to discover a new molecule with great potential as a multi-site fungicide.” The group started by looking for essential cellular processes that were distinctly different between fungi and plants. Using published literature, they identified mitochondrial respiration as a candidate, because fungi have specific respiratory proteins. Previous studies have shown that lipophilic cations accumulate in mitochondria and the team identified the existing fungicide dodine as a potential anti-mitochondrial agent. Using fluorescent fungal reporter strains and live cell imaging, they demonstrated that dodine inhibits mitochondrial respiration, therefore depleting the pathogen of energy. With dodine as the lead compound, they then designed and tested newly synthesised lipophilic cations. One of these novel molecules was found to not only inhibit respiration, but also induce reactive oxygen species, activating the fungal cell death pathway.6 Moreover, this molecule also activated the plant’s immune system, alerting the host to prospective attack. Indeed, this novel molecule has been shown to afford effective protection against fungal diseases such as rice blast and Septoria tritici blotch, most likely due to its multiple activities.
CONTROLLING CANKER WITH CRISPR
With plant pathogens constantly evolving to overcome defences, it can be a struggle to stay one step ahead, especially if the crop in question is a challenge for traditional breeding techniques. Citrus—the world’s most produced fruit—poses considerable challenges for plant breeders, owing to its genetic complexity, prolonged juvenile stage, self- and cross-incompatibility, and pollen and ovule sterility. Consequently, almost all citrus-producing regions are threatened by citrus pathogens, including the devastating citrus canker, caused by the bacterium Xanthomonas citri. “Citrus canker is considered the second most important citrus disease worldwide and affects most citrus-producing countries,” says Nian Wang (University of Florida, USA). “It causes the trees to drop unripe fruits prematurely, affects yields by reducing photosynthesis, and causes lesions to develop on the fruit, making them unmarketable. Without control measures, crop losses may reach up to 80%.” But rapid gene editing technologies such as CRISPR-Cas9 could give producers an advantage.
Canker symptoms develop when Xanthomonas secretes an effector molecule, PthA4, that activates host expression of a susceptibility gene called lateral organ boundaries 1 (LOB1). Nian and his colleagues investigated whether CRISPR can generate canker-resistant citrus varieties by mutating the DNA-binding elements of the LOB1 promoter that PthA4 recognises.7 Their ambition was to generate homozygous resistant plants immediately in the T0 generation, which would allow citrus breeders to bypass the lengthy juvenile stage of these crops to generate homozygous lines in the T1 generation.
To do this, Nian’s group used pummelo, a large citrus fruit popular in Asia that has two identical alleles for LOB1. They first constructed a DNA vector coding for a plant-optimised CRISPR-Cas9 nuclease targeted to the PthA4-binding elements. Young pummelo seedlings were transformed using Agrobacterium infection of the epicotyl region, generating eight transgenic lines. Sequencing revealed that one of these (#Pum1) was a homozygous mutant (with the exact same mutation in both alleles), whilst #Pum6 was a biallelic mutant (with a different mutation in both alleles).
“We then tested the canker resistance of the modified pummelo lines and wild-type pummelo by inoculating them with Xanthomonas citri,” says Nian. “After 5 days, canker symptoms were observed on all of pummelo plants except #Pum1 and #Pum6, which were completely resistant to the disease.” To verify that the canker resistance of #Pum1 and #Pum6 was due to the directed mutation in LOB1, the group infected the plants with X. citri containing a vector coding for a modified PthA4, designed to recognise a different DNA region that could activate LOB1. Because this region was downstream of PthA4’s normal binding element, the modified effector could still activate LOB1 even if the original binding element was mutated. At 5 days post-inoculation, both the wild-type and genome-modified plants showed canker symptoms, demonstrating that the resistant phenotype had been caused by the CRISPR-Cas9 targeted mutation.
Nian and his colleagues intend to use CRISPR to develop canker-resistant citrus varieties that can be used by citrus growers worldwide. “Until now, developing homozygous resistant plants for perennial tree crops has been a challenge, particularly citrus crops which have a long juvenile stage,” says Nian. “But our results demonstrate that CRISPR technology can be used to generate homozygous mutations in the first generation, allowing timely phenotype assessment and therefore more rapid improvement of elite citrus varieties.”
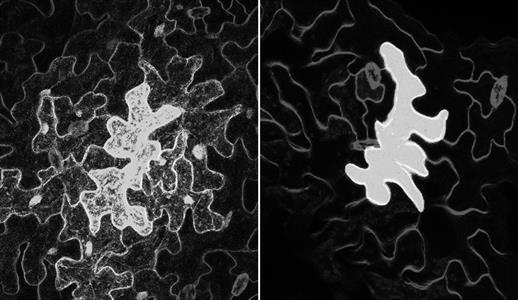
We are now all used to the idea of using ‘lockdowns’ to control the spread of deadly pathogens. It appears that plant cells do the same, by rapidly sealing up the channels that functionally connect cells, known as plasmodesmata. Under normal conditions, water, nutrients, hormones, and other molecules freely pass through these, but during a pathogen’s attack, plant cells respond by rapidly producing the polysaccharide callose. This accumulates at the plasmodesmata and blocks the channels, limiting the spread of virus particles and fungal/bacterial effector molecules that suppress immune responses. Recent research indicates that this process is tightly regulated by specific receptors located within the membrane surrounding the plasmodesmata themselves: the first evidence of these channels initiating an immune response independently to the rest of the cell.
“Although the plasma membrane which lines the plasmodesmata is continuous with that which surrounds the rest of the cell, we now know it is a distinct ‘microdomain’, containing highly specific receptors and lipids,” says Christine Faulkner (John Innes Centre, UK). Christine’s work is starting to uncover the specific signalling machinery that regulates plasmodesmata-related responses, starting with the closure of plasmodesmata in response to chitin, a fungal cell wall compound.8 Using the model plant Arabidopsis, she took a genetic approach that assessed loss-of-function mutants affected in different membrane receptors. “Surprisingly, we found that plasmodesmata closure does not depend on the receptor CERK1 [chitin elicitor receptor kinase 1], which is essential for most chitin-triggered responses in plant cells,” Christine says. Instead, chitin-driven plasmodesmata closure is dependent on the receptor-like protein LysM-containing GPI-anchored protein 2 (LYM2), which localises specifically to plasmodesmata in response to chitin. Here, it appears to form a complex with LysM receptor-like kinase 4 (LYK4) to initiate a downstream signal cascade that induces callose production.
“Ultimately, both the chitin receptors in the plasma membrane and those in the plasmodesmata trigger the production of reactive oxygen species (ROS), but this occurs through distinct downstream signalling components and results in a different outcome,” says Christine. “In the main body of the cell, chitin-induced ROS production is thought to create a pathogen-hostile environment outside the cell, whereas at the plasmodesmata, the result is callose accumulation.” This distinction is clear from the fact that chitin-triggered ROS production in the main body of the cell is unaffected in lym2 mutants. Curiously, both signalling pathways converge on the NADPH oxidase respiratory burst oxidase homolog protein D (RBOHD). Christine’s work has identified that chitin-triggered plasmodesmata closure is dependent on specific phosphorylated serine residues of RBOHD that differ from those required for the CERK1-mediated ROS burst.
“Our work has demonstrated that membrane domain specialisation enables independent control of plasmodesmata in immune signalling,” says Christine. This suggests that cell-to-cell connectivity must be regulated independently of other immune outputs—but why is this the case? “Potentially, this represents a trade-off where the plant cell has to balance the benefits of connectivity with the protection that isolating itself would give to other cells.” Not dissimilar, then, to a government weighing up the benefits of imposing a lockdown during a pandemic with the need to keep essential services functioning.
Interestingly, pathogen effectors may target and counteract plasmodesmata closure: certain fungal effector molecules have been shown to localise to these channels. “For fungi, open plasmodesmata would facilitate both the spread of effector molecules and increased access to host sugars to fuel their growth,” says Christine.
It can be particularly challenging to diagnose plant pathogens when the attack takes place below ground. Nematode worms are highly destructive parasites that infect plant roots, penetrating the host vascular system and disrupting water and nutrient uptake. Collectively, they cause an estimated US $80–$118 billion dollars’ worth of damage per year to crop plants. “One of the major problems with nematodes is that they produce symptoms similar to other diseases or physiological malfunction, such as drought, so farmers try to solve the problem in the wrong way. Particularly in many developing regions, many farmers are not even aware of what nematodes are,” says Ernesto San-Blas (The Centre of Science and Technology for Sustainable Development, La Serena, Chile). Currently, the only way to diagnose nematode infestation is to send soil or root samples for laboratory analysis—a laborious and lengthy process requiring trained expertise.
But Ernesto’s work could open the way for a rapid diagnostic method that could be used even by farmers in the field. His idea is based on Fourier-transform infrared spectroscopy (FTIR), which can identify chemical groups and bonds by measuring the absorption of infrared light by a sample. When an organism’s physiology and metabolism is disrupted by stress events, their molecular composition changes, which then affects the absorption of infrared light. “The concept came completely by chance,” says Ernesto. “Just as I was setting up my lab, my wife, Mayamarú Guerra (Universidad Tenològica de Bolivar, Colombia), a spectroscopy expert, was awarded a grant for an FTIR machine to characterise materials. I asked her what it did and her explanation gave me the idea of seeing whether different species of nematodes produce characteristic spectra.” After finding this to be the case, Ernesto quickly moved on to exploring whether FTIR could identify nematode-infected plants simply by measuring molecular changes in their leaves.
Ernesto and his colleagues infected tomatoes and guava plants with the nematode Meloidogyne enterolobii and took leaf samples each week until the plants showed visible symptoms of infection (at 8 and 12 weeks, respectively).9 Using FTIR, he found that the absorption spectra were significantly different at week 5 post-infection for tomatoes and week 8 for guavas, even though there were no visible symptoms of disease. But he did not stop there. Instead, he decided to investigate whether diagnosis could be accelerated further with artificial intelligence. Using machine learning, a computer was trained to recognise FTIR spectra of healthy and diseased plants from a subset of the data. “When we presented the rest of the experimental data to the computer model, it was able to automatically classify the plants from week 2 in tomatoes and week 4 in guavas, therefore halving the diagnosis time,” says Ernesto.
Ernesto and his colleagues now intend to expand this work by comparing FTIR spectra of plants affected by a wider range of pathogens and also environmental stress. “Our aim is to construct a spectral database to support the development elsewhere of remote sensing technology,” he says. “In the future, we envisage that a farmer will have a portable FTIR device with the installed diagnostic software. They will be able to rapidly take a spectral measurement in the field, and this can immediately tell them whether the crops need water or fertilisers, or if they are being affected by a certain pathogen. Potentially, these devices could even be part of autonomous monitoring robots that send information directly to the farmer’s mobile phone.”
As these examples show, experimental biology is already offering new ways to combat deadly plant diseases. In an increasingly uncertain world, it is likely that we will need to apply all of our knowledge and understanding in increasingly innovative ways against both plant and animal pathogens.
1. Fisher MC, Henk DA, Briggs CJ, et al. 2012. Emerging fungal threats to animal, plant and ecosystem health. Nature 484: 186–194.
2. Bebber DP, Ramotowski MAT, Gurr SJ. 2013. Crop pests and pathogens move poleward in a warming world. Nat Clim Chang 3: 985-988.
3. Chaloner TM, Fones HN, Varma V, et al. 2019. A new mechanistic model of weather-dependent Septoria tritici blotch disease risk. Philos Trans R Soc Lond B Biol Sci 374: 20180266.
4. Fones HN, Bebber DP, Chaloner TM, et al. 2020. Threats to global food security from emerging fungal and oomycete crop pathogens. Nat Food 1: 332–342.
