TPJ September 2023 Editor choice: Where do they come from, where do they go: cell lineage tracing with CRISPR
A versatile CRISPR-based system for lineage tracing in living plants (https://doi.org/10.1111/tpj.16378)
Where do they come from, where do they go: cell lineage tracing with CRISPR
Early studies of cell lineages in plants discovered the clonal nature of the three germ layers, L1, L2, and L3 in the shoot apical meristem. Nowadays most optical cell lineage tracing methods use reporter genes that are expressed in a time-, tissue-, or trigger-dependent manner and encode enzymes such as GUS or lacZ, which convert substrates into colorful products. For the substrate reaction, however, the tissue must be fixed and lineages can therefore not be traced in vivo. Additionally, it is crucial for lineage tracing that only few cells express the reporter gene, so that lineages of single cells can be observed. Mattia Donà and colleagues use nuclear fluorescent proteins that are expressed depending on the mutagenic activity of CRISPR as cell lineage tracing reporters for living tissue. The reporter system labels the nuclei of individual cells based on frameshifts induced by CRISPR/Cas9, with mClover fused to Histone 2B (H2B) as a readout (Figure 1A).
The authors tested their system based on well-described cell lineages in the root epidermis. Root hair cells are derived from trichoblasts, and non-root hair cells from atrichoblasts, and their fate is determined by their contact with either one or two adjacent cortex cell files. The authors used the GLABRA2 (GL2) promoter, which is active during the early differentiation of cells in the root meristem, to drive Cas9 expression with the LT1 or LT2 constructs. They observed labeling in adjacent cell files in the root epidermis, which overlapped with a reporter expressed under the GL2 promoter in the atrichoblast lineage, confirming that once the lineage-tracing reporter is activated, expression remains active despite cell division (Figure 1B). The authors also confirmed the function of the reporter in the shoot apical meristem, where they expressed it under the CLV3 promoter, which is active in all three layers of the central zone, and showed that the cell lineage labeling followed the L1, L2, and L3 structure (Figure 1B).
Although the frequency of frame-shift corrections was high enough to show events in nearly all plants, the authors detected only a few transmissions to the next generation and therefore speculated that only a small number of events were generated in the germline. They crossed LT1 or LT2 constructs with a pCLV3:H2B-mCherry reporter line to obtain F1 progeny with a single copy of the LT construct and then scored the F1 plants for visible fluorescent sectors. Around 70 % of the F1 seedlings had fluorescent sectors, suggesting that the reporter had not been activated in the germline of the parent plant but was still active in the F1 generation. They then analysed F2 seedlings for mClover expression and counted around 80 % of the seedlings with fluorescent sectors. This shows that plants that have lost their reporter due to activity of the CRISPR construct in the germline represent only a very small proportion of the progeny and can be identified at seedling stage and discarded before seed propagation.
To test their reporter in a haploid species, the authors expressed the CRISPR-mClover constructs under Marchantia-specific promoters in Marchantia. LT1 and LT2 constructs under the MpYUC2 promoter formed sectors with fluorescent nuclei, which originated in the meristems at the notches of the gemmae (Figure 1C), suggesting that the reporter system is also functional in Marchantia and can be used for cell lineage tracing studies.
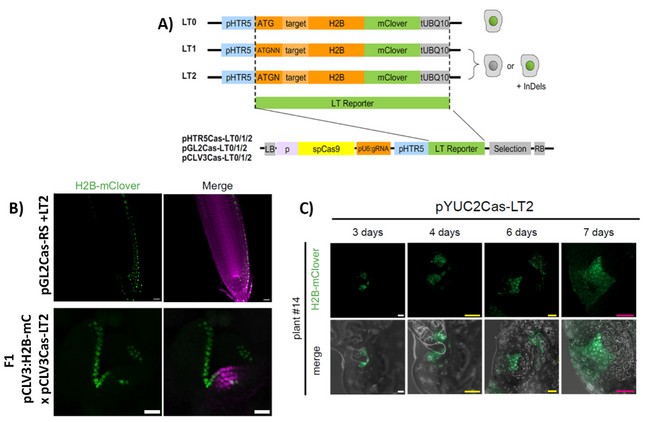
Figure 1: CRISPR-based system to trace cell lineages in Arabidopsis and Marchantia.
A) Scheme of the CRISPR-mClover constructs; mClover is fused to a Histone H2B (H2B) for nuclear localisation under the HRT5 promoter; depending on the construct, the inserted gRNA target close the start codon of mClover leads to a functional open reading frame (L0), or a +1 or +2 frameshift (LT1, LT2).
B) H2B-mClover signal (green) in Arabidopsis root cell lineage induced by Cas9 expressed under the GL2 promoter (upper panel), and in a shoot apical meristem cell lineage induced by Cas9 expressed under the CLV3 promoter (lower panel).
C) H2B-mClover signal (green) in Marchantia gemmae, induced by Cas9 expressed under the YUC2 promoter; figure modified from (Donà et al., 2023).





