JXB Volume 73, Issue 8 – Editor’s choice
This article highlights the following publication:
Custom methods to identify conserved genetic modules applied to novel transcriptomic data from Amborella trichopoda
Ana C Rivarola Sena, Amélie Andres-Robin, Aurelie C Vialette, Jérémy Just, Alexandra Launay-Avon, Néro Borrega, Bertrand Dubreucq, Charles P Scutt
Journal of Experimental Botany, Volume 73, Issue 8, 18 April 2022, Pages 2487–2498, https://doi.org/10.1093/jxb/erac044
Getting to the roots of flowers
The evolution of flowers is a significant milestone in earth’s history since it changed global landscapes and biomes drastically. For millions of years the planet was colonized by ferns, mosses, and trees until the first plants started to blossom, and so did plant biodiversity. The emergence of flowers led to a surge in the number of angiosperm species within a relatively short geological time scale. With over 300,000 different species, angiosperms are the largest and most diverse group within the plant kingdom and comprise most of our cultivated crops. Charles Darwin called the rapid development of higher plants that he found in fossil records an “abominable mystery” because it seemed to contradict his assumption of gradual evolution without major leaps. Demystifying the origin of the angiosperm flower is a central focus of evolutionary developmental biology. Evolution changes species over generations by gradually modulating their embryonic development, and analysing how expression patterns especially of developmental genes change over time can give important insight into the evolutionary success of angiosperms.
In this issue of the Journal of Experimental Botany Rivarola Sena et al. (2022) describe characters of the most recent common ancestor of all flowering plants that existed at least 149 million years ago, and they do so without the need of fossil records. The authors compare transcriptomic data from two extant species, Amborella trichopoda and the model plant Arabidopsis thaliana, whose phylogenetic lineages have diverged at the very beginning of the angiosperm clade. They identify shared characteristics that were likely already present in the common ancestor of all flowering plants and were conserved during angiosperm evolution.
Amborella is a very special plant that hides deep in the understory of the tropical forest of New Caledonia, a remote island group off the East coast of Australia, where it has existed for about 130 million years. This rather unimposing shrubby plant (Figure 1A) is the only living representative of Amborellales, the most basal order of flowering plants that formed the earliest side branch in the angiosperm family tree. Amborella is thus a sister group to all other flowering plants and its genomic sequence is a window into the deep past of angiosperm evolution. Major parts of Amborella’s genome have therefore been sequenced and have been described in parts. However, to address the question of the origin of the angiosperm flower, detailed transcriptomic data of the different flower organs such as ovule, carpel, and stigma are essential not only at maturity but also in developing flower buds, and these transcriptomic resources have until now been limited. Analysing Amborella buds is not straightforward since they are under 1mm in diameter (Figure 1B, C). Rivarola Sena et al. therefore used laser-assisted microdissection to present the first detailed transcriptomic data of Amborella female flower bud components (Figure 1D-F). Interestingly, Amborella is dioecious with female and male flowers on different individuals, whereas the most recent common ancestor of angiosperms likely had bisexual flowers.
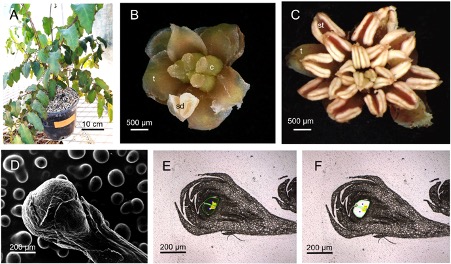
Figure 1: Amborella trichopoda is a shrubby flowering plant endemic to New Caledonia (A). This dioecious species grows small female (B) and male flowers (C) on different individuals. In their study Rivarola Sena et al. present the first detailed transcriptomic data of different organs from Amborella female flower buds (D). The bud components were removed using laser-assisted microdissection (E, F; longitudinal sections before and after removal of carpel tissues). c, carpel; sd, staminode; st, stamen; t, tepal.
To compare Amborella and Arabidopsis gene expression the team used the newly generated transcriptomic data from flower buds and additionally assembled published organ-specific RNA sequencing datasets from both species. A critical step in the analysis and comparison of such large genome-wide expression data is the use of module detection methods to group genes into co-expression sets. For this they applied weighted gene co-expression network analysis to find groups of genes whose expression is highly correlated. This analysis was followed by comparison of gene orthogroups between Amborella and Arabidopsis. These genes are derived from a single gene in the last common ancestor of both species. The authors identified nine gene modules representing over 5,000 orthogroups whose expression profiles have been conserved and have likely been present before the evolutionary radiation of angiosperm species. From these results the authors partially reconstructed the expression patterns of the nine gene modules in bud, flower, root, and leaf tissue of the most recent common ancestor of all angiosperms. Interestingly, around one third of the genes that are key regulators of female development in Arabidopsis are among the identified orthogroups, indicating that their expression patterns and functions have little changed in the course of evolution.
With this study Rivarola Sena et al. prove that comparison of transcriptomic data of species that have large phylogenetic distance can successfully be used to infer conserved traits already present in their common ancestor. Importantly, the team used data generated in independent studies performed by different groups, which shows that published datasets can reliably be used to compare gene expression-clustering data between species and identify conserved gene modules. However, to achieve meaningful results, the authors caution that care should be taken to compare transcriptomic data of similar tissues and developmental stages.
To successfully exploit the exciting possibilities data sharing offers to address macro-evolutionary questions, it is furthermore of importance to adhere to the FAIR principles defined by Wilkonson et al. (2016), that encourage Findability, Accessibility, Interoperability, and Reusability of research results. Only then can the scientific community reuse and build on published data with confidence.
Deducing developmental features of plants that have existed millions of years ago from living specimens provides important insight into the origin of flowers. It remains to be determined, however, where to place this significant event on the geological time scale. Especially the time point that Amborella diverged from all other angiosperms, the so-called ‘crown age’, is controversially debated. Why different dating approaches result in diverging age estimates ranging from 140 – 270 million years can be read in the latest JXB Flowering Newsletter, in which Sauquet et al. (2022) argue that despite considerable research on this topic the angiosperm crown age is still best described as being unknown. Overcoming this uncertainty will be an exciting challenge to be addressed in the future.
References
Sauquet H, Ramírez-Barahona S, Magallón S. 2022. What is the age of flowering plants? Journal of Experimental Botany, erac130, https://doi.org/10.1093/jxb/erac130
Wilkinson M, Dumontier M, Aalbersberg I et al. 2016. The FAIR Guiding Principles for scientific data management and stewardship. Scientific Data 3, 160018. https://doi.org/10.1038/sdata.2016.18





