November 2025 TPJ Editor choice: Coding the carpel: evolutionary roots of NGATHA genes in seed plants
Highlighting: https://doi.org/10.1111/tpj.70488
Coding the carpel: evolutionary roots of NGATHA genes in seed plants
In gymnosperms (‘naked seeds’ in ancient Greek), ovules are exposed, usually on the surface of cone scales. In flowering plants (angiosperms – ‘enclosed seeds’), however, leaves evolved into carpels that folded over the ovules, fully enclosing them inside an ovary. This enclosure protects ovules from drying out, pests, and damage. It also controls which pollen tubes can reach the ovules and allows for the development of complex reproductive strategies such as double fertilization and fruit formation. Evolutionarily, this enclosure was a massive step that allowed plants to evolve more efficient and selective reproduction, leading to the incredible diversity of flowering plants today.
In Arabidopsis, different transcription factor families play crucial roles in regulating carpel development. Although angiosperms appeared relatively suddenly in the fossil record, their evolutionary origins remain unresolved. Gymnosperms represent the only other extant group of seed plants, and interestingly, many genes related to floral organ development have gymnosperm orthologs. To investigate the evolution of gene families essential for carpel formation, Ignacio Cota and colleagues tested whether gymnosperm homologs could substitute for their angiosperm counterparts in Arabidopsis.
NGATHA (NGA) genes in Arabidopsis have a redundant role in the formation of the style and stigma. They belong to the RAV family, characterized by the presence of a B3 DNA-binding domain. In Arabidopsis, there are seven NGA or NGA-related genes, and six other RAV genes, which possess an additional AP2 domain. NGA genes are considered exclusive to angiosperms, whereas RAV genes with an AP2 domain appear in all streptophytes, including streptophyte algae, bryophytes, lycophytes, ferns, and seed plants.
The authors identified five RAVs with an AP2 domain in the gymnosperm Ginkgo. Overexpression of GbRAV5 in Arabidopsis resulted in a phenotype reminiscent of the overexpression of Arabidopsis NGA genes, including dwarfism, fasciated stems, fewer petals, and shorter siliques with a characteristic expansion at the distal end. GbRAV5 overexpression restored the formation of the stigma in Arabidopsis nga1 nga3 double mutants, suggesting that GbRAV5 is functionally equivalent to NGA factors.
The transcription factors HECATE (HEC) and SPATULA (SPT) have key roles in controlling overall growth of the stigma, style and transmitting tract. Overexpression of GbHEC and GbSPT in the respective Arabidopsis mutants complemented the phenotypes in the carpels. Overexpression of CRABS CLAW (CRC), a YABBY (YAB) transcription factor also involved in carpel formation, however, did not complement the Arabidopsis crc mutant phenotype, suggesting that the significant divergence of CRC from other YAB genes might be a key innovation in the evolution of the carpel.
NGA and HEC proteins interact with various partners to perform their functions, including INDEHISCENT (IND) and SPT. In Arabidopsis, the NGA–HEC dimer subsequently forms a tetrameric complex with SPT and IND upon activation, which drives stigma development. The authors used bimolecular fluorescence complementation assays to show that these protein–protein interactions are conserved between Ginkgo and Arabidopsis. In Ginkgo ovules, GbHEC expression overlapped with GbRAV5 expression, suggesting that both genes together could control female reproductive structures in Ginkgo.
Previous studies suggested that NGAs were unique to angiosperms, distinguishing them from other RAV proteins that contain both AP2 and B3 domains. However, GbRAV5 could rescue the nga mutant phenotype in Arabidopsis, challenging this view. To explore this, the authors built a phylogeny of RAV homologs from Ginkgo and ten other gymnosperms. They found that 37 gymnosperm RAV proteins clustered with NGAs rather than with AP2-containing RAVs. Most of these NGA-like gymnosperm proteins showed partially degenerated or missing AP2 domains, making them resemble angiosperm NGAs. Overexpression of RAV5 homologs from three gymnosperms restored style and stigma formation in Arabidopsis nga1 nga3 mutants in two cases, and even GbRAV5 lacking its AP2 domain rescued the phenotype, indicating the AP2 domain is not required for NGA-related function.
The authors highlight that the parallel loss of the AP2 domain offers a rare view of an evolutionary intermediate. Such intermediates are likely to be evolutionarily unstable and short-lived, making the presence of partial AP2 domains in many gymnosperm genes especially noteworthy.
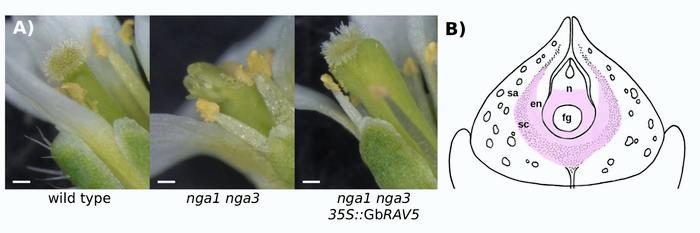
Figure 1: GbRAV5 overexpression complements the nga1 nga3 mutant in Arabidopsis and is expressed with GbHEC in Ginkgo ovules after pollination.
A) Stigmas of wild type, the nga1 nga3 double mutant and when GbRAV5 is overexpressed in the nga1 nga3 double mutant.
B) GbRAV5 and GbHEC are expressed in the Ginkgo ovule after pollination (pink), including in the basal part of the nucellus (n) and the inner layers of the integument that will differentiate into the sclerotesta (sc) and the endotesta (en). Figure modified from (Cota et al., 2025).






