TPJ February 2022 Editor's choice by Leonie Verhage
Temporal changes in transcripts of Miniature Inverted‐Repeat Transposable Elements during rice endosperm development.
Nagata, H., Ono, A., Tonosaki, K., Kawakatsu, T., Sato, Y., Yano, K., Kishima, Y. and Kinoshita, T.
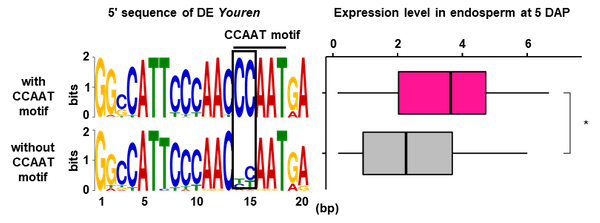
Youren insertions in the genome and transcription factor-binding motifs. Sequence logos for the 5' sequences of Youren with (upper, n = 165) and without (bottom, n = 33) the CCAAT motif. The horizontal line above the logos indicates the conserved CCAAT motif. The box indicates the initial CC of the CCAAT motif, which is not conserved in the minor group of Youren sequences. (c) Box plots of the expression level of Youren sequences with (magenta) and without (gray) the CCAAT motif in endosperm at 5 DAP. *, P-value = 0.05 using a Wilcoxon rank sum test. Figure from Nagata et al., 2022.
Can transposable elements rewire transcriptional networks in the developing rice endosperm?
Transposable elements (TEs) are generally silenced by epigenetic mechanisms, such as DNA methylation, to prevent genome instability and deleterious mutations. However, methylation marks are passively and actively removed from TE sequences during the plant life cycle and in various tissues, including the endosperm. It is not completely clear why TEs are expressed in this tissue. One hypothesis suggests that the transposable elements expressed in the endosperm are processed into small RNAs and transported to the embryo to repress expression of TEs there (as reviewed by Park et al., 2020). Additionally, the endosperm is a terminally differentiated tissue and therefore may tolerate high TE activity.
To understand why TEs are expressed, it is important to characterize their expression patterns. Although genome-wide hypomethylation and TE transposition are well described in the endosperm, knowledge of how TE expression changes over time during seed development is lacking. In issue 109:5 of The Plant Journal, Hiroki Nagata and colleagues at the Yokohama City University in Japan publish their findings after studying the dynamics of TE expression in the endosperm of rice. The rice genome is relatively small among grasses, but contains a high number of TEs. Moreover, TEs in rice populate not only the pericentromeric regions, as in Arabidopsis, but also the euchromatin, making rice is a suitable model for their study.
The rice genome is enriched in TEs of the DNA transposon class, which move to different locations in the genome through a DNA intermediate. Miniature-inverted repeat TEs (MITEs) are especially abundant. MITEs are internal deletion derivatives of autonomous DNA transposons with terminal inverted repeats at both ends. They are typically 100-600 bp in length and mostly reside in the euchromatin relatively close to genes. For this reason, standard short-read RNAseq is not appropriate for measuring TE expression in rice, as during library preparation, short RNAs are lost. Therefore, the researchers constructed a microarray with probes that were designed from annotated TEs in the TIGR Plant Repeat Database.
When they analyzed the expression of endosperm TEs at different time points, the authors noticed that higher expression of TEs mostly takes place at later developmental stages, between 7 and 10 days after pollination. Moreover, MITEs were prominent amongst the differentially expressed TEs. The researchers found differential expression of several types of MITE families, but one type caught their attention: Youren, a MITE that belongs to the Tourist-like category of elements. Many of the differentially expressed Youren elements from different positions within the rice genome showed similar expression behavior, with the highest expression values at 5 to 6 days after pollination. This comparable behavior of Youren transcripts from different positions within the rice genome suggests that their transcription is in some way coordinated. This was supported by the finding that Youren elements are transcribed from the DNA in only one direction.
To understand the basis for directional transcription, the scanned all Youren copies for cis-regulatory elements and found a CCAAT motif, which is the typical binding site for NF-Y transcription factors. As the authors detected expression of several NF-Y transcription factors in the embryo and the endosperm, they reasoned that the NF-Y binding motif is a good candidate to explain the directional transcription of Youren sequences. Moreover, when they compared the expression levels of different Youren copies, they found that the expression levels of the copies with the motif were higher than of the copies without the motif (Figure).
Tetsu Kinoshita, the corresponding author, is especially interested in the discovery of the NF-Y binding sites. Some members of the NF-Y transcription factor family, such as LEC1, are known as pioneer transcription factors. Pioneer factors can bind to their target binding sites in both ‘open’ and ‘closed’ chromatin and initiate changes to the local chromatin structure. In this way, they can either open or close the neighboring chromatin and regulate the expression of the corresponding genes. Hence, the NF-Y binding site in Youren elements could be involved in rewiring transcriptional networks.
Leonie Verhage, Research Highlights Editor





