May 2025 TPJ Editor choice: Twists in the pattern: REM transcription factors determine phyllotaxis in the Arabidopsis inflorescence
Highlighted publication:
Behind phyllotaxis, within the meristem: a REM-ARF complex shapes inflorescence in Arabidopsis thaliana
https://doi.org/10.1111/tpj.70041
Twists in the pattern: REM transcription factors determine phyllotaxis in the Arabidopsis inflorescence
Patterns that form with mathematical precision are remarkably prevalent in nature. The spirals seen in the scales of a pinecone or the seeds of a sunflower are examples of phyllotaxis, a self-organising process through which lateral organs arise at constant divergence angles around a central axis. These patterns are based on the Fibonacci sequence, where each number is the sum of the preceding two (1, 1, 2, 3, 5, 8, 13, 21, …). Dividing a full circle of 360° by the ratio of two consecutive Fibonacci numbers results in a larger arc of 222.5° and a smaller arc of 137.5°. This “golden angle” of 137.5° is precisely the angle by which leaf or flower primordia are separated from one another. Why this pattern is so prevalent in nature remains unclear; it has been suggested it might improve light capture, maximise organ packing, or be due to developmental constraints within the meristem.
At the tissue level, phyllotactic patterns at the shoot apical and inflorescence meristems are shaped by gradients of the plant hormone auxin. Auxin is initially produced at the center of the meristem and transported to regions at the meristem’s periphery where a new leaf or flower primordium will form. The primordium depletes the surrounding area of auxin and thereby creates an “inhibitory field” that defines the spacing between adjacent organs. Yet, while auxin’s role in establishing phyllotaxis is well understood, additional regulators of this process remain to be identified.
Caselli and colleagues investigated a putative role for Arabidopsis REPRODUCTIVE MERISTEM 34 (REM34) and REM35 in this process. These transcription factors are predominantly expressed during inflorescence development, but further analysis has been hampered by the lack of mutant alleles for REM35 and by the tight linkage of the two genes on chromosome 4 that effectively prevents the generation of double mutants by crossing. Caselli and colleagues overcame this obstacle using CRISPR/Cas9 technology. Analysis of the newly generated CRISPR mutants showed that the phyllotactic pattern of siliques was altered in rem34 and rem35, with fewer siliques displaying the golden angle of 137.5°. This phenotype was exacerbated in the rem34 rem35 double mutant, with more angles falling into 80-100° or 170-190° intervals (Fig. 1A). These observations suggest that the two REM genes jointly regulate phyllotaxis.
Caselli et al. then used a yeast-2-hybrid interaction screen to identify proteins that act in concert with REM34 and REM35. They identified AUXIN RESPONSE FACTOR 19 (ARF19) as an interactor of REM35, but not of REM34, and the same interaction pattern was observed for the closely related protein ARF7. ARF7 and ARF19 act redundantly in lateral root development but had no previously described role in inflorescence development. However, phyllotaxis was similarly perturbed in arf7 arf19 and rem34 rem35 double mutants. Fluorescent reporter lines showed that all four genes are expressed in the inflorescence meristem, with their expression domains overlapping in the L1 and L2 layers, and in the boundary regions between the meristem and young flower primordia. Overall, these results suggest that REM-ARF protein complexes act in the inflorescence meristem to position new flower primordia.
Two known ARF7/ARF19 target genes, PUCHI and LATERAL BOUNDARY DOMAIN 18 (LBD18), are also expressed in the inflorescence meristem. The promoters of both genes were found to be directly bound by REM34 and REM35 and their expression pattern at the shoot apex was strongly perturbed in arf7 arf19 and rem34 rem35 mutants. Loss-of-function mutants of LBD18 and PUCHI displayed altered phyllotaxis similar to that of rem34 rem35 and arf7 arf19, corroborating the hypothesis that these two genes act downstream of REM-ARF complexes in the control of inflorescence meristem architecture.
Taken together, this study provides first evidence for a REM-ARF transcription factor complex that regulates inflorescence architecture. While REM34, REM35, ARF7 and ARF19 transcripts are present in the inflorescence meristem, their expression patterns only partially overlap, and their function might depend on the presence or absence of their interaction partners. The authors propose that different REM-ARF dimers or trimers form simultaneously within distinct meristem domains and fulfil different roles to finetune boundary determination and, consequently, primordia positioning.
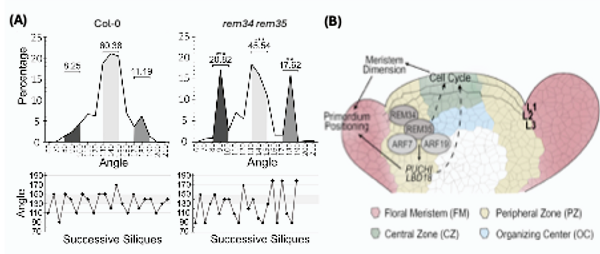
Figure 1. REM34 and REM35 control phyllotactic patterns in the inflorescence meristem
- Upper panels show the phyllotactic pattern of wild type (Col-0) and rem34 rem35 mutant plants. Lower panels display the phyllotactic pattern of a single representative plant.
- Model of REM function in the Arabidopsis inflorescence meristem.
Figure modified from Caselli et al. (2025).





