TPJ article: Dynamically expressed small RNAs, substantially driven by genomic structural variants, contribute to transcriptomic changes during tomato domestication
Small RNAs, big changes – how sRNAs might have driven tomato domestication
Qing, Y., Zheng, Y., Mlotshwa, S., Smith, H. N., Wang, X., Zhai, X., Knaap, E., Wang, Y., Fei, Z.
Dynamically expressed small RNAs, substantially driven by genomic structural variants, contribute to transcriptomic changes during tomato domestication
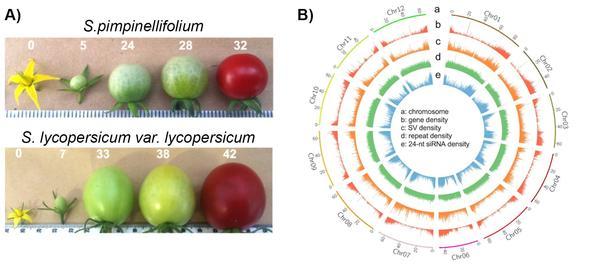
Like all crop plants, modern tomato (Solanum lycopersicum) has undergone a long domestication process, which can be traced back to wild, red-fruited progenitors 80.000 years ago in the Andean region. When tomatoes arrived in Europe in the 15th or 16th century, domestication forged ahead, as breeders selected for fruit traits like larger fruits and enhanced flavor, modified growth habits like self-pruning and reduced plant height, and increased resistance to biotic and abiotic stresses. Tomato is now one of the most important fruit crop in terms of yield worldwide.
Recent research suggests that genomic structural variants (SVs) are strongly associated with phenotypic changes favored by the breeding process, and that multiple SVs control the dosage and expression of genes that modify fruit flavor, size, and production. SVs can cause large changes in cis-regulatory regions and thereby alter gene expression, or possibly even alter gene copy number directly. However, a molecular link between gene expression changes and SVs is often missing.
Small RNAs (sRNAs) in plants also play a major role in gene regulation. MicroRNAs (miRNAs) and phased secondary short interfering RNAs (phasiRNAs) are 21-22 nt RNAs generated by Dicer-like enzymes that are loaded into the RNA-induced silencing complex (RISC) in order to execute their function: binding a target mRNA and facilitating its degradation. In contrast, 24-nt heterochromatic small interfering RNAs (hc-siRNAs) guide RNA-directed DNA methylation to confer epigenetic regulation on gene expression. The role of sRNAs in macroevolution, such as the evolution from algae to flowering plants, has been studied in depth, but their role in short-term evolutionary processes such as domestication, has not yet been analyzed.
In issue 110:6 of The Plant Journal, Qing and colleagues investigated the link between SVs and sRNAs. They selected nine tomato accessions, ranging from wild tomato, to semi-domesticated and fully domesticated and analysed their transcriptomes, including sRNAs. They found that all three classes of sRNAs showed significant changes in expression based on their domestication history, and that these changes were linked to underlying SVs. Specifically, hc-siRNA abundance was strongly correlated with the density of SVs. Additionally, for those hc-siRNAs that mapped to SVs, a higher proportion of them were differentially expressed. Given that hc-siRNAs are involved in DNA methylation, this suggests that SVs might play a strong role for epigenetics during domestication. The domestication-associated epigenetic changes were often related to biotic and abiotic stress responses in addition to several developmental processes, but the most statistically significant ones were related to pathogen resistance. Qing and colleagues discovered that SVs contributed to changes in phasiRNA-generating loci, which also primarily mapped to disease resistance genes.
Overall, SV-related dynamic expression changes in sRNAs mostly regulate stress response and growth genes, suggesting that tomato domestication primarily targeted these processes. A similar prioritization of stress response genes might have happened during domestication of other crops. Some of the sRNA loci, such as SV overlapping hc-siRNAs, could be used as targets for breeding, especially for improving biotic and abiotic stress tolerance in modern tomatoes. The knowledge of sRNA loci and SVs that influence these traits can be used by breeders to identify useful alleles in the wild tomato germplasm and create improved genotypes by introgressing and pyramiding unused natural alleles.





